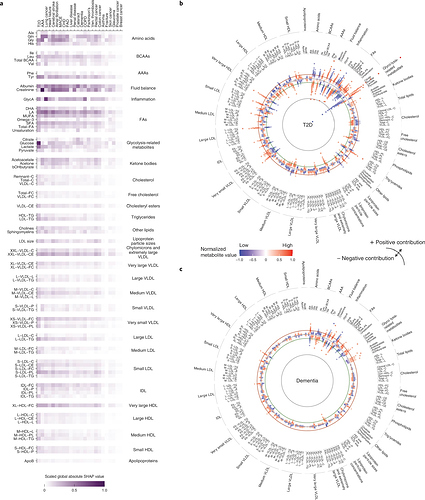
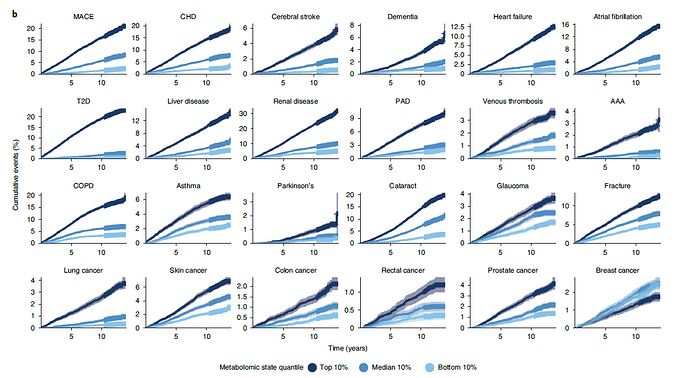
That’s an intense study.
I think they used one source for the training, and other sources for validation:
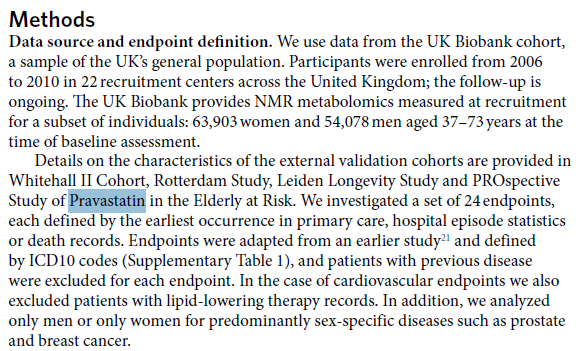
I think the Pravastatin study was used for validation, not training.
For a neural network (NN), for training, they should just put data into it and set the output to be whatever they are looking for. For instance, if it’s a NN to find cat pictures, they input different pictures, and the output will be whether or not the input picture is a picture of a cat. That’s often in the form of a probability, eg, “there’s a 90% chance the picture is of a cat”. They keep doing this until the NN meets some set of criteria indicating it’s as good as it’s going to get.
Typically, what’s done next is they use a separate set of pictures to see how well the NN actually does.
And though I’ve used the example of a single output, you could have multiple outputs: cat; dog; horse; person; etc. These may be likelihoods, so that you’d have to see the outputs to determine which entity is supposed to be the most likely.
I think that’s what they did here: they used the UK Biobank cohort for training; then did validation using other “cohorts”. In other words, using other datasets, how well does our model (NN) actually perform at showing who will get what disease?
They say it’s good, of course. 
It will take a lot more reading and deciphering to really analyze the paper, though. Sadly, time I don’t have.